Rows: 1,236
Columns: 8
$ case <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 1…
$ bwt <int> 120, 113, 128, 123, 108, 136, 138, 132, 120, 143, 140, 144, …
$ gestation <int> 284, 282, 279, NA, 282, 286, 244, 245, 289, 299, 351, 282, 2…
$ parity <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ age <int> 27, 33, 28, 36, 23, 25, 33, 23, 25, 30, 27, 32, 23, 36, 30, …
$ height <int> 62, 64, 64, 69, 67, 62, 62, 65, 62, 66, 68, 64, 63, 61, 63, …
$ weight <int> 100, 135, 115, 190, 125, 93, 178, 140, 125, 136, 120, 124, 1…
$ smoke <int> 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 0, 1, …Simple Linear Regression
Data babies in openintro package
Baby Weights
Baby Weights
lm stands for linear model
se stands for standard error
| y | Response | Birth weight | Numeric |
|---|---|---|---|
| x | Explanatory | Gestation | Numeric |
Linear Equations Review
Recall from your previous math classes
\(y = mx + b\)
where \(m\) is the slope and \(b\) is the y-intercept
e.g. \(y = 2x -1\)

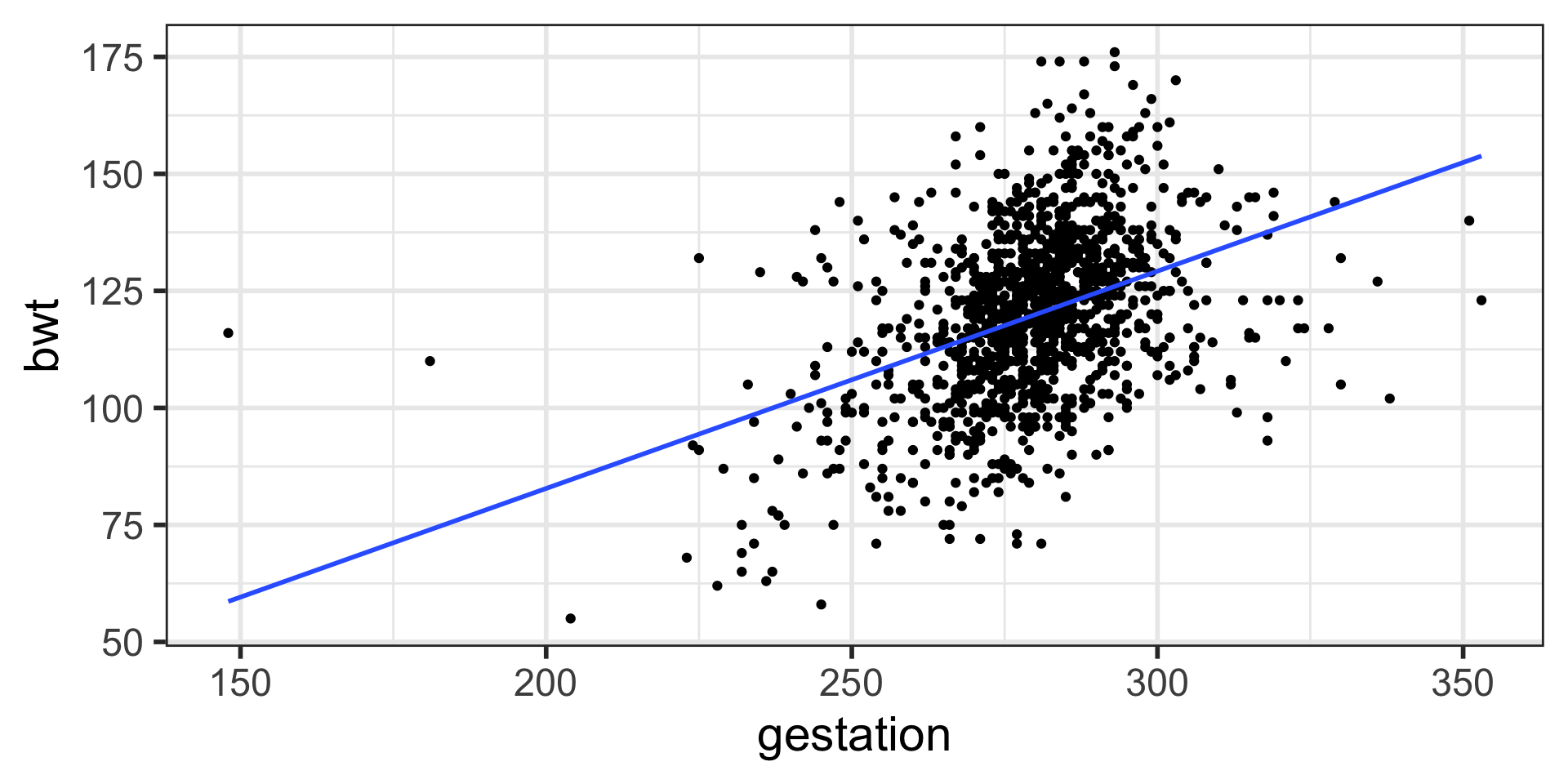
Notice anything different between baby weights plot and this one?
Math class
\(y = b + mx\)
\(b\) is y-intercept
\(m\) is slope
Stats class
\(y_i = \beta_0 +\beta_1x_i + \epsilon_i\)
\(\beta_0\) is y-intercept
\(\beta_1\) is slope
\(\epsilon_i\) is error/residual
\(i = 1, 2, ...n\) identifier for each point
Notation
| Sample Statistic | Population Parameter | |
|---|---|---|
| Intercept | \(b_0\) | \(\beta_0\) |
| Slope | \(b_1\) | \(\beta_1\) |
| Error/Residual | \(e\) | \(\epsilon\) |
lm stands for linear model. We are fitting a linear regression model. Note that the variables are entered in y ~ x order.
# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -10.1 8.32 -1.21 2.27e- 1
2 gestation 0.464 0.0297 15.6 3.22e-50\(\hat {y}_i = b_0 + b_1 x_i\)
\(\hat {\text{bwt}_i} = b_0 + b_1 \text{ gestation}_i\)
\(\hat {\text{bwt}_i} = -10.1 + 0.464\text{ gestation}_i\)
Expected bwt for a baby with 300 days of gestation
\(\hat {\text{bwt}_i} = -10.1 + 0.464\text{ gestation}_i\)
\(\hat {\text{bwt}} = -10.1 + 0.464 \times 300\)
\(\hat {\text{bwt}} =\) 129.1
For a baby with 300 days of gestation the expected birth weight is 129.1 ounces.
Interpretation of estimates

\(b_1 = 0.464\) which means for one unit(day) increase in gestation period the expected increase in birth weight is 0.464 ounces.

\(b_0 = -10.1\) which means for gestation period of 0 days the expected birth weight is -10.1 ounces!!!!!!!! (does NOT make sense)
Extrapolation
- There is no such thing as 0 days of gestation.
- Birth weight cannot possibly be -10.1 ounces.
- Extrapolation happens when we use a model outside the range of the x-values that are observed. After all, we cannot really know how the model behaves (e.g. may be non-linear) outside of the scope of what we have observed.
Baby number 148

Baby #148
Expected
\(\hat y_{148} = b_0 +b_1x_{148}\)
\(\hat y_{148} = -10.1 + 0.464\times300\)
\(\hat y_{148}\) = 129.1
Observed
\(y_{148} =\) 160
Residual for i = 148

\(y_{148} = 160\)
\(\hat y_{148}\) = 129.1
\(e_{148} = y_{148} - \hat y_{148}\)
\(e_{148} =\) 30.9
Least Squares Regression
The goal is to minimize
\[e_1^2 + e_2^2 + ... + e_n^2\]
which can be rewritten as
\[\sum_{i = 1}^n e_i^2\]
Conditions
Conditions for Least Squares Regression
Linearity
Normality of Residuals
Constant Variance
Independence
Linear

Non-linear

Nearly normal

Not normal

Constant Variance

Non-constant variance

Independence
Harder to check because we need to know how the data were collected.
In the description of the dataset it says [a study]considered all pregnancies between 1960 and 1967 among women in the Kaiser Foundation Health Plan in the San Francisco East Bay area.
It is possible that babies born in the same hospital may have similar birth weight.
Correlated data examples: patients within hospitals, students within schools, people within neighborhoods, time-series data.
Inference
Inference: Hypothesis Testing
\(H_o: \beta_1 = 0\)
\(H_A: \beta_1 \neq 0\)
# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -10.1 8.32 -1.21 2.27e- 1
2 gestation 0.464 0.0297 15.6 3.22e-50Since the p-value of 3.22e-50 < 0.05 we reject the null hypothesis and conclude that there is a significant relationship between gestation and birth weight.
Inference: Confidence Interval
CI = point estimate \(\pm\) critical value \(\times\) standard error
95% CI for the slope = point estimate of the slope \(\pm\) critical value \(\times\) standard error of the slope
Critical value
Inference: Confidence Interval
# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) -10.1 8.32 -1.21 2.27e- 1
2 gestation 0.464 0.0297 15.6 3.22e-5095% CI = 0.4642626 \(\pm\) 1.9618867 \(\times\) 0.0297437
95% CI = (0.4059089, 0.5226163)
Inference: Confidence Interval
Inference: Confidence Interval
Note that the 95% confidence interval for the slope does not contain zero and all the values in the interval are positive indicating a significant positive relationship between gestation and birth weight.
Confidence Interval
| y | Response | Birth weight | Numeric |
|---|---|---|---|
| x | Explanatory | Smoke | Categorical |
Notation
\(y_i = \beta_0 +\beta_1x_i + \epsilon_i\)
\(\beta_0\) is y-intercept
\(\beta_1\) is slope
\(\epsilon_i\) is error/residual
\(i = 1, 2, ...n\) identifier for each point
# A tibble: 2 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 123. 0.649 190. 0
2 smoke -8.94 1.03 -8.65 1.55e-17\(\hat {y}_i = b_0 + b_1 x_i\)
\(\hat {\text{bwt}_i} = b_0 + b_1 \text{ smoke}_i\)
\(\hat {\text{bwt}_i} = 123 + (-8.94\text{ smoke}_i)\)
Expected bwt for a baby with a non-smoker mother
\(\hat {\text{bwt}_i} = 123 + (-8.94\text{ smoke}_i)\)
\(\hat {\text{bwt}_i} = 123 + (-8.94\times 0)\)
\(\hat {\text{bwt}_i} = 123\)
\(E[bwt_i | smoke_i = 0] = b_0\)
Expected bwt for a baby with a smoker mother
\(\hat {\text{bwt}_i} = 123 + (-8.94\text{ smoke}_i)\)
\(\hat {\text{bwt}_i} = 123 + (-8.94\times 1)\)
\(\hat {\text{bwt}_i} = 114.06\)
\(E[bwt_i | smoke_i = 1] = b_0 + b_1\)
Note that the confidence interval for the “slope” does not contain 0 and all the values in the interval are negative.
Understanding Relationships
Just because we observe a significant relationship between \(x\) and \(y\), it does not mean that \(x\) causes \(y\).
Just because we observe a significant relationship in a sample that does not mean the findings will generalize to the population.
For these we need to understand sampling and study design.